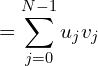Religion is regarded by the common people as true, by the wise as false, and by the rulers as useful.
Lucius Annaeus Seneca

Dalle’s Idea of Religion
First as always i hope everyone is safe oh Dear Readers. Secondly, i am going to write about something that i have been pondering for quite some time, probably close to two decades.
What i call Religion Of Warez (ROWZ).
This involves someone who i hold in the highest regard YOU the esteemed developer.
Marc Andreeson famously said “Software Is Eating The Word”. Here is the original blog:
“Why Software Is Eating The World” by Marc Andreeson
There is a war going on for your attention. There is a war going on for your thumb typing. There is a war going on for your viewership. There is a war going on for your selfies. There is a war going on for your emoticons. There is a war going on for github pull requests.
There is a war going on for the output of your prompting.
We have entered into The Great Cognitive Artificial Intelligence Arms Race (TGCAIAR) via camps of Large Languge Model foundational model creators.
The ability to deploy the warez needed to wage war on YOU Oh Dear Reader is much more complex from an ideological perspective. i speculate that Software if i may use that term as an entity is a non-theistic religion. Even within the Main Tabernacle of Software (MTOS) there are various fissures of said religions whether it be languages, architectures or processes.
A great blog can be found here -> Software Development: It’s a Religion.
Let us head over to the LazyWebTM and do a cursory search and see what we find[1] concerning some comparison numbers for religions and software languages.
In going to wikipedia we find:
According to some estimates, there are roughly 4,200 religions, churches, denominations, religious bodies, faith groups, tribes, cultures, movements, ultimate concerns, which at some point in the future will be countless.
Wikipedia
Worldwide, more than eight-in-ten people identify with a religious group. i suppose even though we don’t like to be categorized, we like to be categorized as belonging to a particular sect. Here is a telling graphic:
Let us map this to just computer languages. Just how many computer languages are there? i guessed 6000 in aggregate. There are about 700 main programming languages, including esoteric coding languages. From what i can ascertain some only list notable languages add up to 245 languages. Another list called HOPL, which claims to include every programming language ever to exist, puts the total number of programming languages at 8,945.
So i wasn’t that far off.
Why so much kerfuffle on languages? For those that have ever had a language discussion, did it feel like you were discussing religion? Hmmmm?
Hey, my language does automatic heap management. Why are you messing with memory allocation via this dumb thing called pointers?
The Art of Computer Programming is mapping an idea into a binary computational translation (classical computing rates apply). This process is highly inefficient compared to having binary-to-binary discussions[2]. Note we are not even considering architectures or methods in this mapping. Let us keep it at English to binary representation. What is the dimensionality reduction for that mapping? What is lost in translation?
For reference, i found a very precise and well-written blog here -> How Much Code Has Ever Been Written?
The calculation involves the number of lines of code ever written up to that point sans the exponential rate from the past two years:
2,781,000,000,000
Roughly 2.8 Trillion Lines of Code have been written in the past 20 years.
Sage McEnery 2020
As i always like to do i refer to Miriam Webster Dictionary. It holds a soft spot in my heart strings as i used to read it in grade school. (Yes i read the dictionary…)
Religion: Noun
re·li·gion (ruh·li·jen)
: a cause, principle, or system of beliefs held to with ardor and faith
Well, now, Dear Reader, the proverbial plot thickens. A System of Beliefs held to faith. Nowadays, religion is utilized as a concept today applied for a genus of social formations that includes several members, a type of which there are many tokens or facets.
If this is, in fact, the case, I will venture to say that Software could be considered a Religion.
One must then ask? Is there “a model” to the madness? Do we go the route of the core religions? Would we dare say the Belief System Of The Warez[3] be included as a prominent religion?

I have said several times and will continue to say that Software is one of the greatest human endeavors of all time. It is at the essence of ideas incarnate.
It has been said that if you adopt the science, you adopt the ideology. Such hatred or fear of science has always been justified in the name of some ideology or other.
If we take this as the undertone for many new aspects of software, we see that the continuum of mind varies within the perception of the universe by which we are affected by said software. It is extremely canonical and first order.
Most often, we anthropomorphize most things and our software is no exception. It is as though it were an entity or even a thing in the most straightforward cases. It is, in fact, neither. It is just information imputed upon our minds via probabilistic models via non convex optimization methods. It is as if it was a Rorschach test that allowed many people to project their own meaning onto it (sound familiar?).
Let me say this a different way. With the advent of ChatGPT we seem to desire IT to be alive or reason somehow someway yet we don’t want it to turn into the terminator.
Stock market predictions – YES
Terminator – NO.
The Thou Shalts Will Kill You
~ Joseph Campbell
Now we are entering a time very quickly where we have “agentic” based large language models that can be scripted for specific tasks and then chained together to perform multiple tasks.
Now we have large language models distilling information gleaned from other LLMs. Who’s peanut butter is in the chocolate? Is there a limit of growth here for information? Asymptotic token computation if you will?
We are nowhere near the end of writing the Religion Of Warez (ROWZ) sacred texts compared to the Bible, Sutras, Vedas, the Upanishads, and the Bhagavad Gita, Quran, Agamas, Torah, Tao Te Ching or Avesta, even the Satanic Bible. My apologies if i left your special tome out it wasn’t on purpose. i could have listed thousands. BTW for reference there is even a religion called the Partridge Family Temple. The cult’s members believe the characters are archetypal gods and goddesses.
In fact we have just begun to author the Religion Of Warez (ROWZ) sacred text. The next chapters are going be accelerated and written via generative adversarial networks, stable fusion and reinforcement learning transformer technologies.
Which, then, one must ask which Diety are YOU going to choose?
i wrote a little stupid python script to show relationships of coding languages based on dates for the main ones. Simple key value stuff. All hail the gods K&R for creating C.
import networkx as nx
import matplotlib.pyplot as plt
def create_language_graph():
G = nx.DiGraph()
# Nodes (Programming languages with their release years)
languages = {
"Fortran": 1957, "Lisp": 1958, "COBOL": 1959, "ALGOL": 1960,
"C": 1972, "Smalltalk": 1972, "Prolog": 1972, "ML": 1973,
"Pascal": 1970, "Scheme": 1975, "Ada": 1980, "C++": 1983,
"Objective-C": 1984, "Perl": 1987, "Haskell": 1990, "Python": 1991,
"Ruby": 1995, "Java": 1995, "JavaScript": 1995, "PHP": 1995,
"C#": 2000, "Scala": 2003, "Go": 2009, "Rust": 2010,
"Common Lisp": 1984
}
# Adding nodes
for lang, year in languages.items():
G.add_node(lang, year=year)
# Directed edges (influences between languages)
edges = [
("Fortran", "C"), ("Lisp", "Scheme"), ("Lisp", "Common Lisp"),
("ALGOL", "Pascal"), ("ALGOL", "C"), ("C", "C++"), ("C", "Objective-C"),
("C", "Go"), ("C", "Rust"), ("Smalltalk", "Objective-C"),
("C++", "Java"), ("C++", "C#"), ("ML", "Haskell"), ("ML", "Scala"),
("Scheme", "JavaScript"), ("Perl", "PHP"), ("Python", "Ruby"),
("Python", "Go"), ("Java", "Scala"), ("Java", "C#"), ("JavaScript", "Rust")
]
# Adding edges
G.add_edges_from(edges)
return G
def visualize_graph(G):
plt.figure(figsize=(12, 8))
pos = nx.spring_layout(G, seed=42)
years = nx.get_node_attributes(G, 'year')
# Color nodes based on their release year
node_colors = [plt.cm.viridis((years[node] - 1950) / 70) for node in G.nodes]
nx.draw(G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=10, font_weight='bold', arrows=True)
plt.title("Programming Language Influence Graph")
plt.show()
if __name__ == "__main__":
G = create_language_graph()
visualize_graph(G)

Programming Relationship Diagram
So, folks, let me know what you think. I am considering authoring a much longer paper comparing behaviors, taxonomies and the relationship between religions and software.
i would like to know if you think this would be a worthwhile piece?
Until Then,
#iwishyouwater <- Banzai Pipeline January 2023. Amazing.
@tctjr
MUZAK TO BLOG BY: Baroque Ensemble Of Vienna – “Classical Legends of Baroque”. i truly believe i was born in the wrong century when i listen to this level of music. Candidly J.S. Bach is by far my favorite composer going back to when i was in 3rd grade. BRAVO! Stupdendum Perficientur!
[1] Ever notice that searching is not finding? i prefer finding. Someone needs to trademark “Finding Not Searching” The same vein as catching ain’t fishing.
[2] Great paper from OpenAI on just this subject: two agents having a discussion (via reinforcement learning) : https://openai.com/blog/learning-to-communicate/ (more technical paper click HERE)
[3] For a great read i refer you to the The Ware Tetralogy by Rudy Rucker: Software (1982), Wetware (1988), Freeware (1997), Realware (2000)
[4] When the words “software” and “engineering” were first put together [Naur and Randell 1968] it was not clear exactly what the marriage of the two into the newly minted term really meant. Some people understood that the term would probably come to be defined by what our community did and what the world made of it. Since those days in the late 1960’s a spectrum of research and practice has been collected under the term.




![Rendered by QuickLaTeX.com \[\alpha = \min \left(1, \frac{\pi(x^) q(x_t | x^)}{\pi(x_t) q(x^* | x_t)} \right)\]](https://www.tedtanner.org/wp-content/ql-cache/quicklatex.com-d0021c453163124b132e82a38dbf20bb_l3.png)
![Rendered by QuickLaTeX.com \[\alpha = \min \left(1, \frac{\pi(x^*)}{\pi(x_t)} \right)\]](https://www.tedtanner.org/wp-content/ql-cache/quicklatex.com-52d53f2c5d02dc0c354ef060a927b5de_l3.png)





 𝕋𝕖𝕕 ℂ. 𝕋𝕒𝕟𝕟𝕖𝕣 𝕁𝕣. (@tctjr)
𝕋𝕖𝕕 ℂ. 𝕋𝕒𝕟𝕟𝕖𝕣 𝕁𝕣. (@tctjr) 
![Rendered by QuickLaTeX.com \[F(x) &= f\f[k] &= \sum_{j=0}^{N-1} x[j]\left(e^{-2\pi i k/N}\right)^j\0 &\leq k < N\]](https://www.tedtanner.org/wp-content/ql-cache/quicklatex.com-1824d6e88be0eea8520d0874ddd199c6_l3.png)